A growing number of people suffer from colorectal cancer,
which is one of the most common cancers. It is essential to diagnose and
treat the cancer as early as possible. The disease may change the microorganism
communities in the gut, and it could be an efficient method to employ gut
microorganisms to predict colorectal cancer. In this study, we selected operational
taxonomic units that include several kinds of microorganisms to predict
colorectal cancer. To find the most important microorganisms and obtain
the best prediction performance, we explore effective feature selection methods.
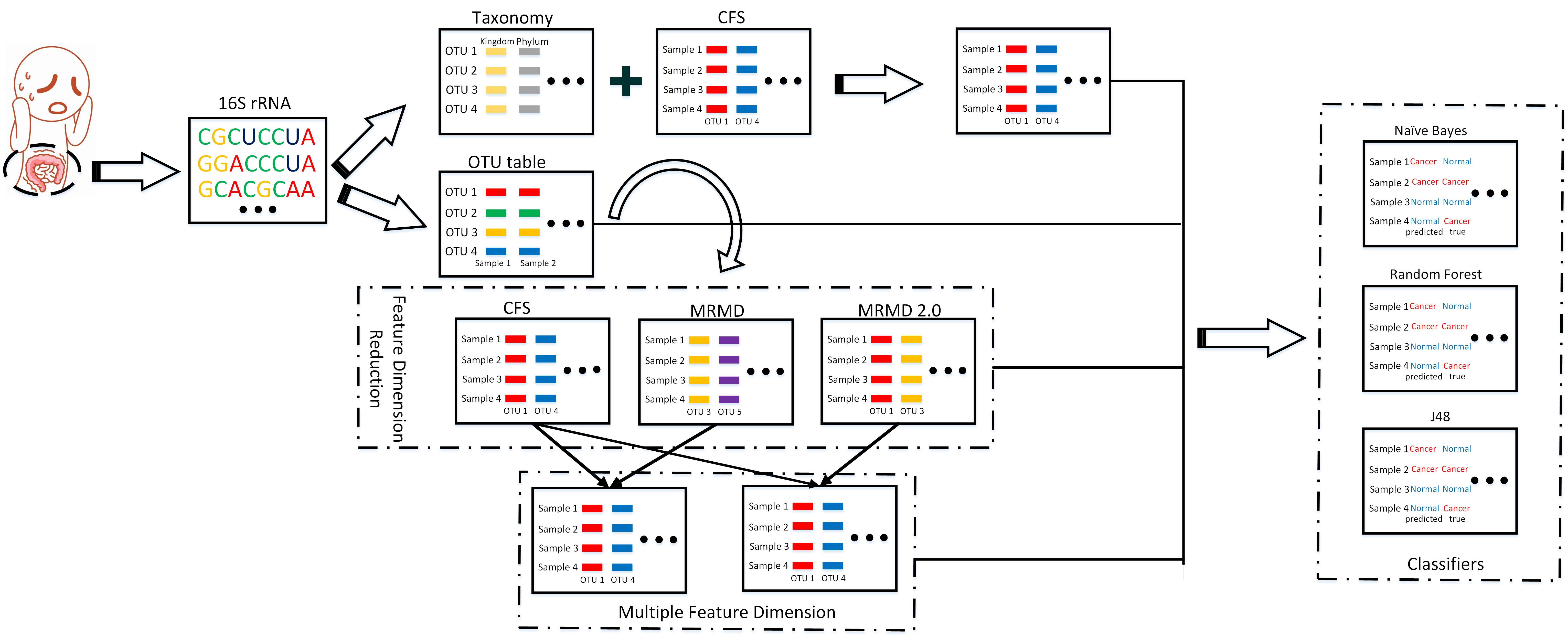
We employ three main steps. First, we use a single method to reduce features.
Next, to reduce the number of features, we integrate the dimension reduction methods
correlation-based feature selection and maximum relevance–maximum distance (MRMD 1.0 and MRMD 2.0).
Then, we selected the important features according to the taxonomy files.
Random forest, naïve Bayes, and decision tree classifiers were evaluated.
Flow Chart
Dataset
Two datasets are used
in this study, both from Oudah et al. The first dataset (CRC1) contains 90 cancer samples and
92 normal samples. The original feature set contains 18,170 OTUs. The second dataset (CRC2)
includes 30 cancer samples and 30 normal samples, with 6,807 OTUs. The study also provides
the taxonomy files, which provide the complete classification information for each OTU
(including kingdom, phylum, class, order, family, genus, and species).
Usage
MRMD 2.0: https://github.com/heshida01/MRMD2.0
CFS: You can find this method in WEKA, and the description is here
Download