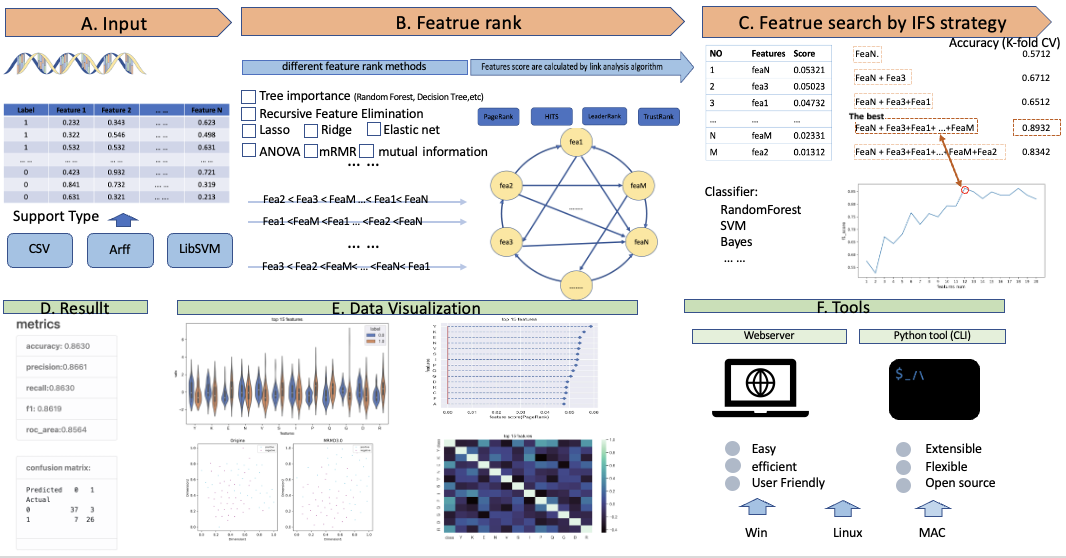
In bioinformatics research, the dimension of data after feature extraction may be very high. As the
number
of features increases, the model will become very complicated, which can easily lead to overfitting.
Although the dimensionality reduction method mentioned above can solve the problem of excessively high
data
dimensions to a certain extent, it requires people to constantly test different feature sizes.
In our previous work, we developed the MRMD1.0 and MRMD2.0 softwares for feature ranking and dimension
reduction.
Here, we propose a new version called MRMD3.0. It is a method that integrates multiple feature ranking algorithms,Which has the following advantages:
Here, we propose a new version called MRMD3.0. It is a method that integrates multiple feature ranking algorithms,Which has the following advantages:
- • built-in multiple feature ranking algorithms
- • four feature link Analysis Algorithms.
- • automatically infer proper dimensions size
- • generate five charts for data analysis
- • VProvides built-in feature selection method's interface for users
- • MRMD3.0 can reduce the dimension of user-specified feature intervals
- ... ...
Feature Rank Method
note:WebServer currently only has built-in integration strategy, the following single method can use our offline command version- ANOVA
- Variance Threshold
- Chisqure
- Linear model emthod ( 1. lasso, 2. ridge, 3.elasticnet)
- mutual inforation( 1.MI 2.NMI 3.MIC)
- minimum Redundancy - Maximum Relevance
- Max-Relevance-Max-Distance
- Recursive Feature Elimination(1. LogisticRegression, 2.SVM, 3.DecisionTreeClassifier)
- tree_feature_importance(1. DecisionTreeClassifier, 2. RandomForestClassifier, 3. GradientBoostingClassifier 4.ExtraTreesClassifier)
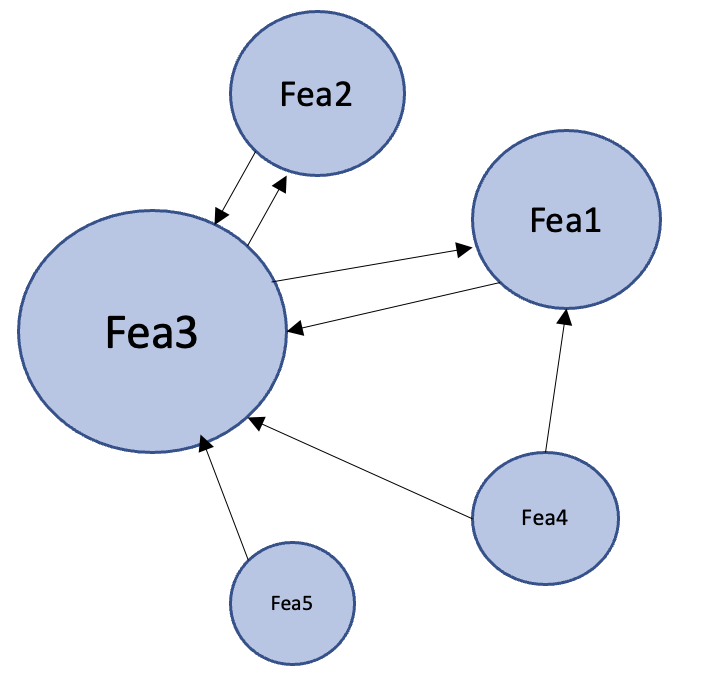
Link Analysis Strategy
- PageRank
- TrustRank
- LeaderRank
- HITS (Authority and Hub)

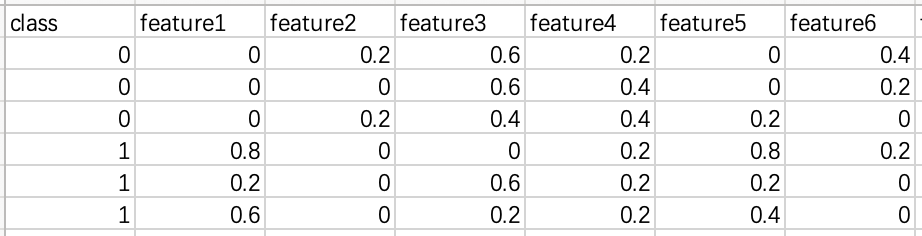
The supported file formats are :
- csv
- arff
- libsvm
Usage Example
python3 mrmd3.0.py [-h] [-s S] -i I [-e E] [-l L] [-n N] [-t T] [-c
{RandomForest,SVM,Bayes}] [-o
O] [-p P] [-m M] [-j J] [-f F] [-r {PageRank,Hits_a,Hits_h,LeaderRank,TrustRank}]
Example
python3 mrmd3.0.py -i test.csv -o out.csv
| # | parameters | description |
|---|---|---|
| • | -s, --start | start feature index, default=1 |
| • | -i, --inputfile | input file (require:arff ,csv or libsvm format) -e, --end |
| • | -e, --end/td> | end index, default=-1 |
| • | -l, --length | step length, default=1 |
| • | -n, --n_dim | mrmd3.0 features top n,default=-1 |
| • | -t, --type_metric | evaluation metric, default=f1 |
| • | -c,--classifier | cross vaildaion classifier r {RandomForest,SVM,Bayes}, default=RandomForest |
| • | -m, metrics_file | output the metrics file's name |
| • | -o, --outfile | output the dimensionality reduction file's name |
| • | -f,--topn | select top n features to chart |
| • | --rank_method | the rank method for features,choices=["PageRank","Hits_a","Hits_h","LeaderRank","TrustRank"],default="PageRank" |