
code: DOWNLOAD
Run nvidia-smi command to see whether the GPU driver is installed properly.
Environments: install anaconda3 and pytorch(Version 1.2 ) first and a Nvidia RTX20XX or RTX1XXX series GPU is required.
<1> conda create -n eFeature python=3.7
<2> conda activate eFeature
<3> conda install pytorch==1.2.0 torchvision==0.4.0 cudatoolkit=10.0 -c pytorch
<4> pip install lightgbm==3.1.1 umap_learn==0.5.1 seaborn==0.11.1 tape_proteins==0.4 imbalanced_learn==0.8.0 joblib==1.0.1 rich==9.12.4 biovec
<5> download eFeature.zip
tar -xvf eFeature.zip
cd eFeature
python eFeature.py -h
The eFeature is configured!!

| Parameters | Description | Notes |
|---|---|---|
| --inTrain | proteins or piptides sequences for TRAINING in FASTA format | FASTA formate seq example:>sequence id(digits or letters)|Label(eg. 0 for negative or 1 for positive) E.G. >seq01|1 MATQAAATTYVVV |
--inTest | proteins or piptides sequences for TESTING in FASTA format | FASTA formate seq example:>sequence id(digits or letters)|Label(eg. 0 for negative or 1 for positive) E.G. >seq01|1 MATQAAATTYVVV |
| --out | output file name in CSV file | The outputs are six deep representations learning features in their separated CSV files and a merged long-feature CSV file of the fused features The 1st column of the CSV file is the sequence id, The 2nd column is the sequence label(0,1,2,...), The columns starting from 3rd columns are feature values(XX_F1,XX_F2,...,XX is the feature name) |
--smote | 0 is without smote or 1 is for doing smote if the Training dataset is imbalanced | Synthetic Minority Oversampling Technique(SMOTE) | --mode | mode=0: only generate features and do feature selection; mode=1: do feature extraction and selection then do classification |
Users could just input FASTA sequence to yield sequence features via mode 0; they could also use mode 1 to generate featers and use six five default machine learning for classification KNN: K Neighbors Classifier LR: Logistic Regression GNB: Gaussian Naive Bayes SVM: support vector machine RF: random forest LGBM: linght gradient boosting machine |
--numclass | the default value is 2 for binary classification; if number of classes is not 2, please specify the exact value. | Do binary classification and it could also do multiclass classification | --kfold | the default values is 5 | Maching learning model trainind via k-fold validation evalution |
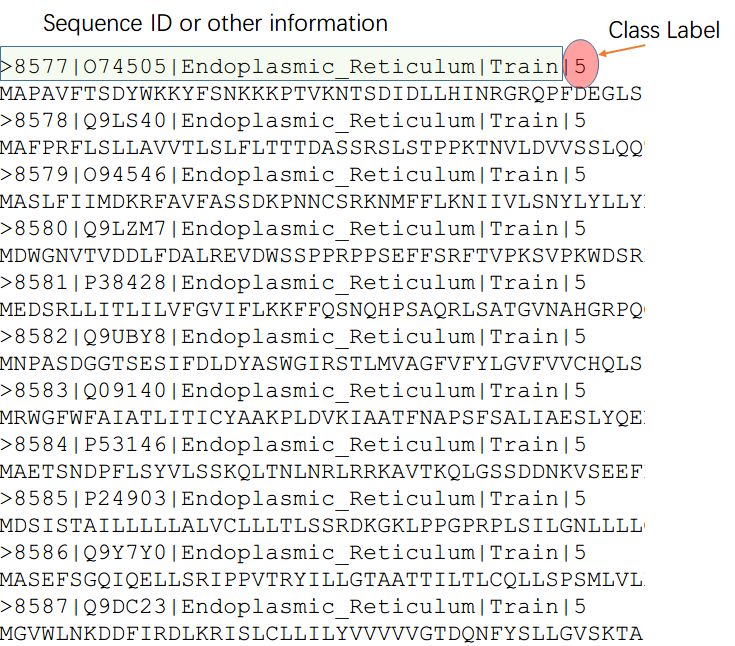
(1)training dataset name: train_data.fasta
(2)test dataset name: test_data.fasta
(2)output file name: subloc.csv
python eFeature.py --inTrain train_data.fasta --out subloc.csv
python eFeature.py --inTrain train_data.fasta --inTest test_data.fasta -out subloc.csv
python eFeature.py --inTrain train_data.fasta --inTest test_data.fasta -out subloc.csv --smote 1
python eFeature.py --inTrain train_data.fasta --inTest test_data.fasta -out subloc.csv --smote 1 --mode 1
python eFeature.py --inTrain train_data.fasta --inTest test_data.fasta -out subloc.csv --smote 1 --mode 1 --kfold 10
python eFeature.py --inTrain train_data.fasta --inTest test_data.fasta -out subloc.csv --mode 2 --numclass 10
| Feature name abbreviations | pretained model |
|---|---|
| lm | Language Model |
| BiLSTM | Bidirectional long short term memory |
| SSA | soft sequence alignment |
| BERT | Bert-based model |
| UniRep | mLSTM | W2V | word2vec |
| FusedAll | The above features are concated to for a long vector |
The Example output of script:
python eFeature.py --inTrain SubLocTrain.fasta --inTest SubLocTest.fasta --out subloc.csv --mode 2 --smote 1 --numclass 10 --kfold 5
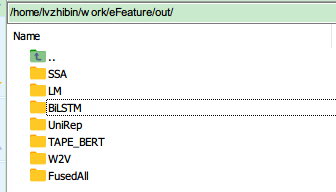
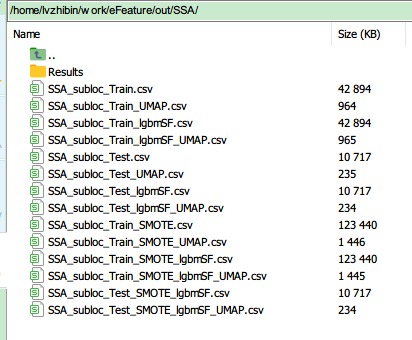
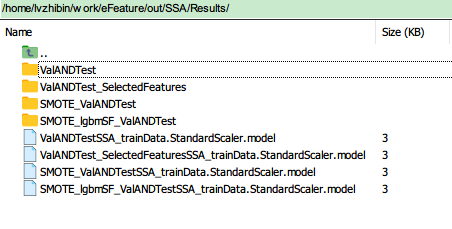
| Foder Name or File name | Meaning |
|---|---|
| ValANDTest | k-fold validation and independent testing results |
| ValANDTest_SelectedFeatures | k-fold validation and independent testing results after feature selection preprocessing |
| SMOTE_ValANDTest | do SMOTE first and get the k-fold validation and independent testing results |
| SMOTE_lgbmSF_ValANDTest | do SMOTE first and follow by a light graident boosting feature selection, then get the k-fold validation and independent testing results |
| Foder Name or File name | Meaning |
|---|---|
| Validation_MeanResults | the mean values of k-fold validation metrics |
| Test_MeanResults | The mean value of independent testing metrics |
| EachFold_Validaiton_Test_Results | The validation and independent testing for each k-fold validated machine-learing model |
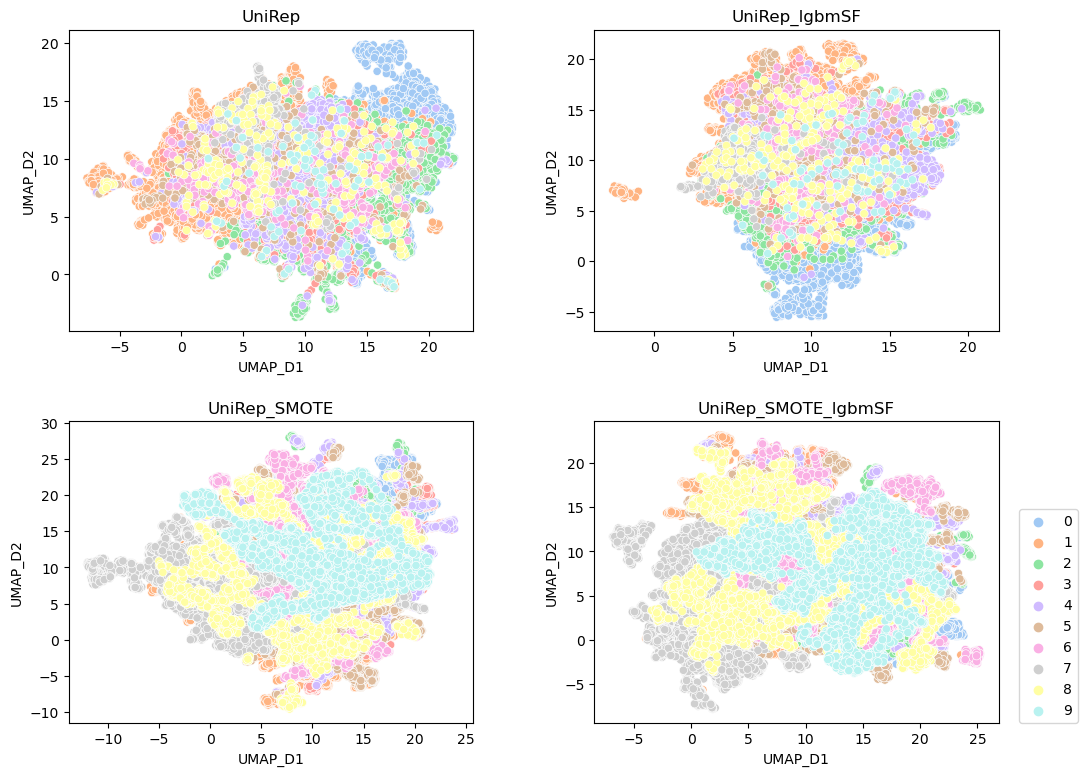
Once the eFeatue value is generated, the UAMP will be done and result will be saved csv file name with UAMP.
run script: python plot.py and you will attain all UAMP data plotting in the plotting folder.

contact: lvzhibin at pku.edu.cn