
DESCRIPTION
|
Maize
(Zea mays) is one of the most
important crops, acting as a source of food, animal feed, and renewable
resources, so improvement of its yield and quality is a major objective. Molecular
biology technologies and multiple omics have received increasing attention
for maize breeding as they provide new and more efficient selection criteria.
In our research, we combined four omics technologies, namely, genomics,
proteomics, transcriptomics of maize, overcoming one technology may address
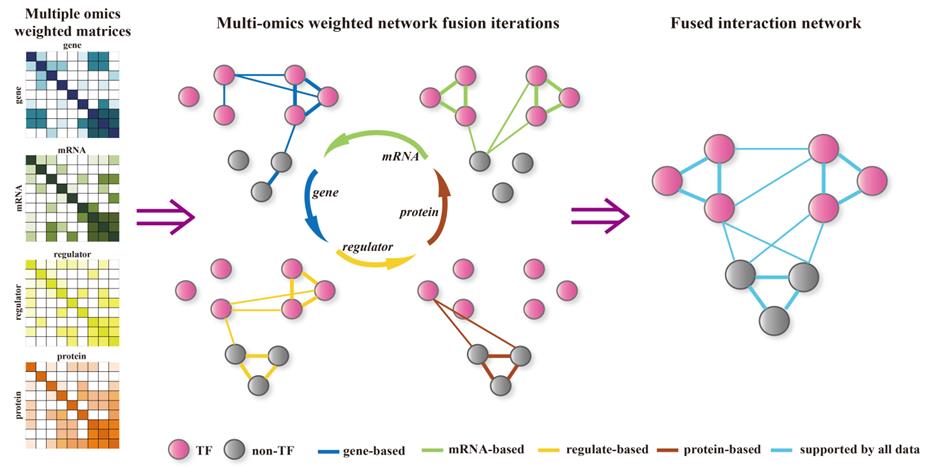
the shortcomings of another help to improve the predicted accurate. A
nonlinear combination method was used to construct networks for each data
type and to integrate these networks into a single similarity network. Here,
we integrated four different weighted networks into a fused network using the
nodes that appeared at least one network. The fused network captures both
shared and complementary information from the different data sources,
offering insight into how informative each data type is for the observed
similarity. After the fusion process, the orphan nodes were taken into the
final fused network, some of which were proved to be TFs that play key roles
in maize. |
DATA ![]()
|
Type |
Description |
Download |
|
Gene interaction network |
We obtained the gene to gene
interaction network from Walley et al’s research [1].
In the first two columns are the interact pairs of genes, and the last column
is the score between them. |
|
|
mRNA interaction network |
We obtained the mRNA to mRNA
interaction network from Walley et al’s research [1].
In the first two columns are the interact pairs of mRNAs, and the last column
is the score between them. |
|
|
Transcriptional regulation network |
We obtained the regulator to target
transcript interaction network from Walley et al’s research [1].
The first column is the regulator, the second column is target transcript and
the last column is the score between them. |
|
|
Protein interaction network |
The protein to protein network comes
from the STRING database because it contains the most known and potential
interact associated among proteins. |
CITATION:
The iterative process of integrating multi-omics weighted networks into a single network
|
Contact |
|
|
Quan
Zou School
of Computer Science and Technology, Tianjin University |
Xiangxiang
Zeng School
of Information Science and Engineering, Xiamen University |
Copyright
© 2018, Xiamen University and Tianjin University Units