
Mengting Niu
I am a associate research fellow at Quan Zou's Lab,the University of Electronic Science and Technology of China,Chengdu, where I work on deep learning, bioinformatic, and computer vision, etc.
Research
Papers:Bioinformatics 2025 (code&data)) Xue Zhang, Quan Zou, Mengting Niu, Chunyu Wang, Predicting circRNA-disease associations with shared units and multi-channel attention mechanisms, Bioinformatics, Volume 41, Issue 3, March 2025, btaf088, https://doi.org/10.1093/bioinformatics/btaf088. (cite) BMC Biology(under review) BMC Biology 2024 (code&data)) Mengting Niu, Chunyu Wang, Yaojia Chen, Quan Zou, Ren Qi, Lei Xu. CircRNA identification and feature interpretability analysis[J]. BMC Biology, 2024, 22(1):44. Doi: 10.1186/s12915-023-01804-x. (cite)
BMC Biology 2024 (code&data)) Mengting Niu, Chunyu Wang, Zhanguo Zhang, Quan Zou. A computational model of circRNA-associated diseases based on a graph neural network: prediction and case studies for follow-up experimental validation[J]. BMC Biology, 2024, 22(1): 24. Doi.org/10.1186/s12915-024-01826-z. (cite)
Briefings in Bioinformatics 2024 Mengting Niu, Chunyu Wang, Yaojia Chen, Quan Zou, Lei Xu. Identification, characterization and expression analysis of circRNA encoded by SARS-CoV-1 and SARS-CoV-2[J]. Briefings in Bioinformatics, 2024, 25(2): bbad537. (cite)
IEEE/ACM Transactions on Computational Biology and Bioinformatics(under review)
Front. Comput. Sci. Mengting Niu,Yaojia Chen, Chunyu Wang, , Quan Zou, Lei Xu. Computational approaches for circRNA-disease association prediction: a review.Frontiers of Computer Science,2025, 19(4): 194904.
PLOS Computational Biology 2022 (code&data)) Mengting Niu, Quan Zou, Chen Lin. CRBPDL: identification of circRNA-RBP interaction sites using an ensemble neural network approach[J]. PLoS Computational Biology, 2022, 18(1): e1009798-e1009798. (cite)
Bioinformatics 2022 (code&data) Mengting Niu, Quan Zou, Chunyu Wang, GMNN2CD: identification of circRNA¨Cdisease associations based on variational inference and graph markov neural networks[J]. Bioinformatics, 2022, 38(8): 2246-2253.(cite)
Briefings in Bioinformatics 2021 Mengting Ni, Ying Ju, Chen Lin, Quan Zou. Characterizing viral circRNAs and their application in identifying circRNAs in viruses[J]. Briefings in Bioinformatics, 2022, 23(1): bbab404. (cite)
IEEE Journal of Biomedical and Health Informatics(data and codes) Mengting Niu, Jin Wu#, Quan Zou, Zhendong Liu, Lei Xu. rBPDL: predicting RNA-binding proteins using deep learning[J]. IEEE Journal of Biomedical and Health Informatics, 2021, 25(9): 3668-3676. (cite)
Plant molecular biology 2021 (data and code ) Mengting Niu, Yuan Lin, Quan Zou. sgRNACNN: identifying sgRNA on-target activity in four crops using ensembles of convolutional neural networks[J]. Plant Molecular Biology, 2021, 105(4-5): 483-495. (cite)
IEEE/ACM Transactions on Computational Biology and Bioinformatics 2021 (web)and dataset) Mengting Niu, Quan Zou. SgRNA-RF: identification of SgRNA on-target activity with imbalanced datasets[J]. IEEE-ACM Transactions on Computational Biology and Bioinformatics, 2021, 19(4): 2442-2453. (cite)
Computational and structural biotechnology journal 2020 (web) and dataset) Niu M, Zhang J, Li Y, et al. CirRNAPL: a web server for the identification of circRNA based on extreme learning machine[J]. Computational and structural biotechnology journal, 2020, 18: 834-842. (cite)
International Conference on Intelligent Computing 2022 Niu M, Hesham A E L, Zou Q. GATSDCD: Prediction of circRNA-Disease Associations Based on Singular Value Decomposition and Graph Attention Network[C]//International Conference on Intelligent Computing. Springer, Cham, 2022: 14-27.(cite)
Journal of proteome research 2019 (web)and dataset) Li Y, Niu M, Zou Q. ELM-MHC: an improved MHC identification method with extreme learning machine algorithm[J]. Journal of proteome research, 2019, 18(3): 1392-1401. (cite)
Recent ProjectsThe main project involved is the related research of circular RNA. Currently, I am leading the GMNN and MSRN project, which aims to develop a series of deep learning models to solve computational problems of circRNA. 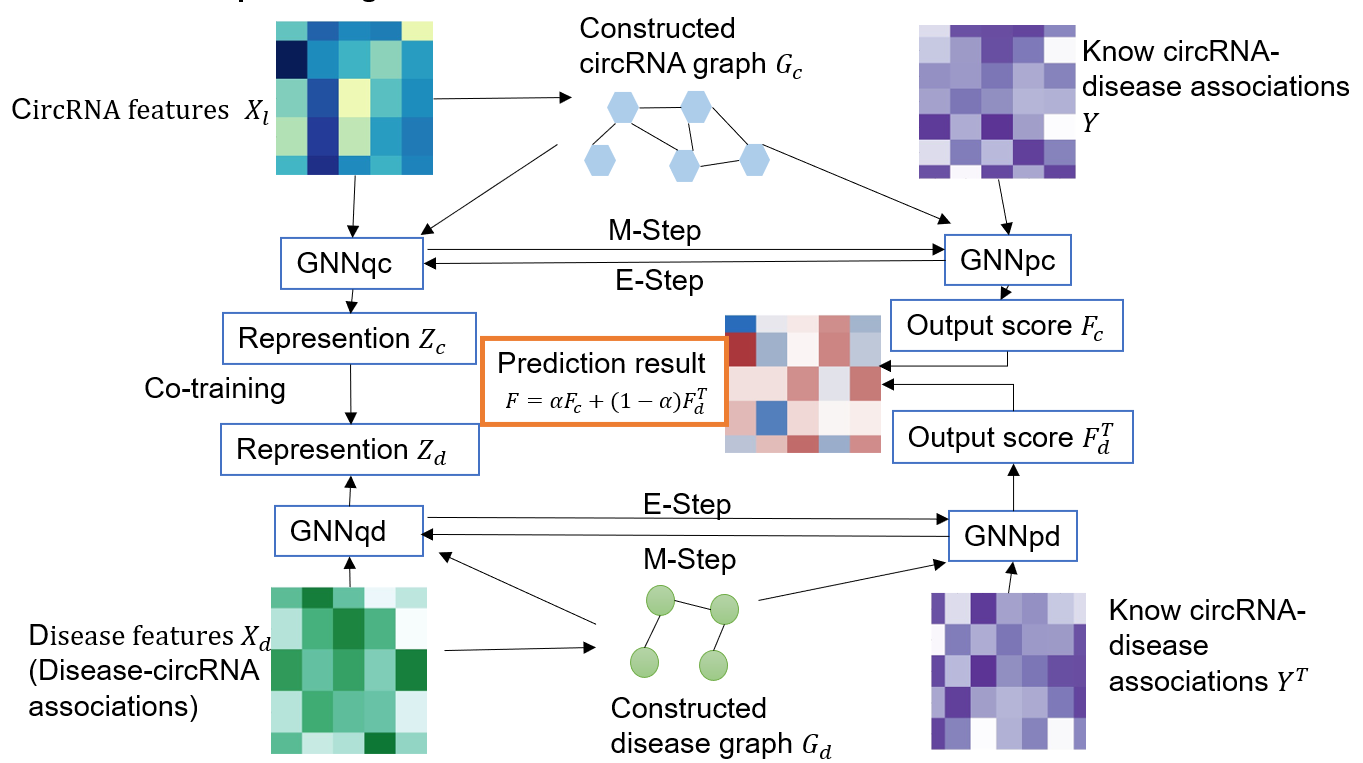

Host.The Natural Science Foundation of China (No.62473268) Host.The Natural Science Foundation of China (No.62303328) Host.The National funded postdoctoral researcher program of China (GZC20230382) Participate: The Natural Science Foundation of China (No.62131004,62172076) Awards
Welcome/Last revision date:2025.3.27
|