
Objective
The identification and analysis the genes of crop yield traits are an essential but challenging task in bioinformatics research. We have developed a database of rice yield and identified candidates based on the database.
People
- Xiangxiang Zeng (Professor, HNU, China)
- Quan Zou (Professor, UESTC, China)
- Chunyu Wang (Associated Professor, HIT, China)
- Jing Jiang (Ph.D, XMU, China)
Publications and Web Servers
- Yansu Wang, Murong Zhou, Quan Zou, Lei Xu. Machine learning for phytopathology: from the molecular scale towards the network scale. Briefings in Bioinformatics. Accepted
- Zheng Chen, Zijie Shen, Da Zhao, Lei Xu, Lijun Zhang, Quan Zou. Genome-wide analysis of LysM-containing gene family in Wheat: evolution and duplications during development and denfences. Genes. 2020, 12(1):31
- Zhibin Lv, Hui Ding, Lei Wang, Quan Zou*. A Convolutional Neural Network Using Dinucleotide One-hot Encoder for identifying DNA N6-Methyladenine Sites in the Rice Genome. Neurocomputing. 2021,422:214-221 (web server)
- Zheng Chen, Zijie Shen, Lei Xu, Da Zhao, Quan Zou*. Regulator network analysis of rice and maize yield- related genes. Frontiers in Cell and Developmental Biology. 2020, 8: 621464
- Mengting Niu, Yuan Lin*, Quan Zou*. SgRNACNN: identifying sgRNA on-target activity in four crops using ensembles of convolutional neural networks. Plant Molecular Biology. DOI: 10.1007/s11103-020-01102-y
- Zijie Shen, Yuan Lin*, Quan Zou*. Transcription factors-DNA interactions in rice: identification and verification. Briefings in Bioinformatics. 2020, 21(3): 946-956 (SCI, IF2018=9.101, PMID:31091308)
- Sanwen Sun, Chunyu Wang, Hui Ding, Quan Zou*. Machine learning and its applications in plant molecular studies. Briefings in Functional Genomics. 2020, 19(1): 40-48 (SCI, IF2018=3.133)
- Changli Feng, Quan Zou*, Donghua Wang*. Using a Low Correlation High Orthogonality Feature Set and Machine Learning Methods to Identify Plant Pentatricopeptide Repeat Coding Gene/Protein. Neurocomputing. Doi: 10.1016/j.neucom.2020.02.079 (SCI, IF2018=4.072)
- Qianfei Huang, Jun Zhang, Leyi Wei, Fei Guo*, Quan Zou*. 6mA-RicePred: A method for identifying DNA N6-methyladenine sites in the rice genome based on feature fusion. Frontiers in Plant Science. 2020, 11: 4 (data) (SCI, IF2018=4.106)
- Jiang, J., F. Xing, C. Wang, and X.Zeng*. "Identification and Analysis of Rice Yield-Related Candidate Genes by Walking on the Functional Network." Frontiers in Plant Science 2018, 9:1685. (SCI, IF2018=4.106, PMID: 30524460) [web server] (EndNote)
- Jiang, J., F. Xing, X. Zeng, and Q. Zou*. "RicyerDB: A Database for Collecting Rice Yield-Related Genes with Biological Analysis." International Journal of Biological Sciences 14, no. 8 (2018): 965-70. (SCI, IF2018= 4.067, PMID: 29989091) [web server] (EndNote)
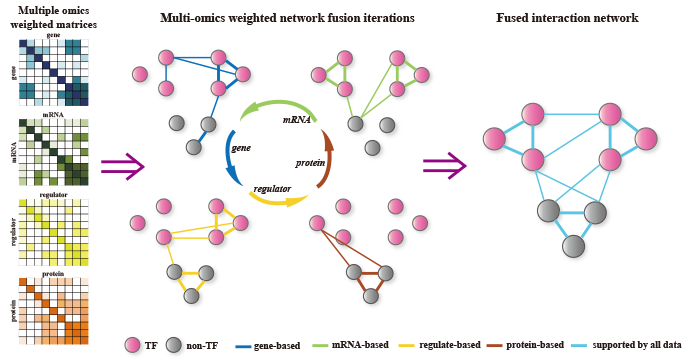
- Jiang, J., F. Xing, X. Zeng, and Q. Zou*. "Investigating maize yield-related genes in multiple omics interaction network data." IEEE Trans Nanobioscience. (2019). (SCI, IF= 1.927, PMID: 31170079) (EndNote)
- Jiang, J., F. Xing, C. Wang, X. Zeng, and Q. Zou*. "Investigation and development of maize fused network analysis with multi-omics." Plant Physiol Biochem. (2019): 380-387. (SCI, IF= 3.404, PMID: 31220804) [web server] (EndNote)
 |
 |
 Friendly web servers were developed and maintained.
Friendly web servers were developed and maintained.
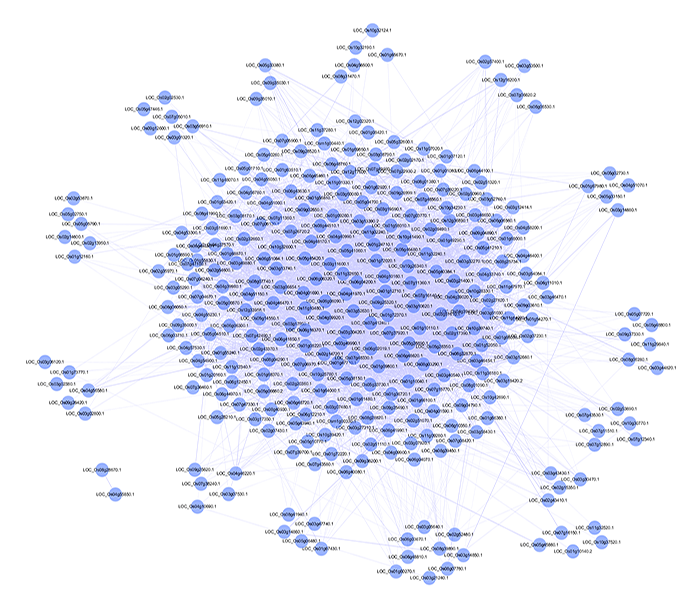
 We manually searched the related databases and literature.
We manually searched the related databases and literature. We proposed a computational systems biology approach for
the identification of candidate genes via a random walk model on a
PPI network with functional similarities.
We proposed a computational systems biology approach for
the identification of candidate genes via a random walk model on a
PPI network with functional similarities.